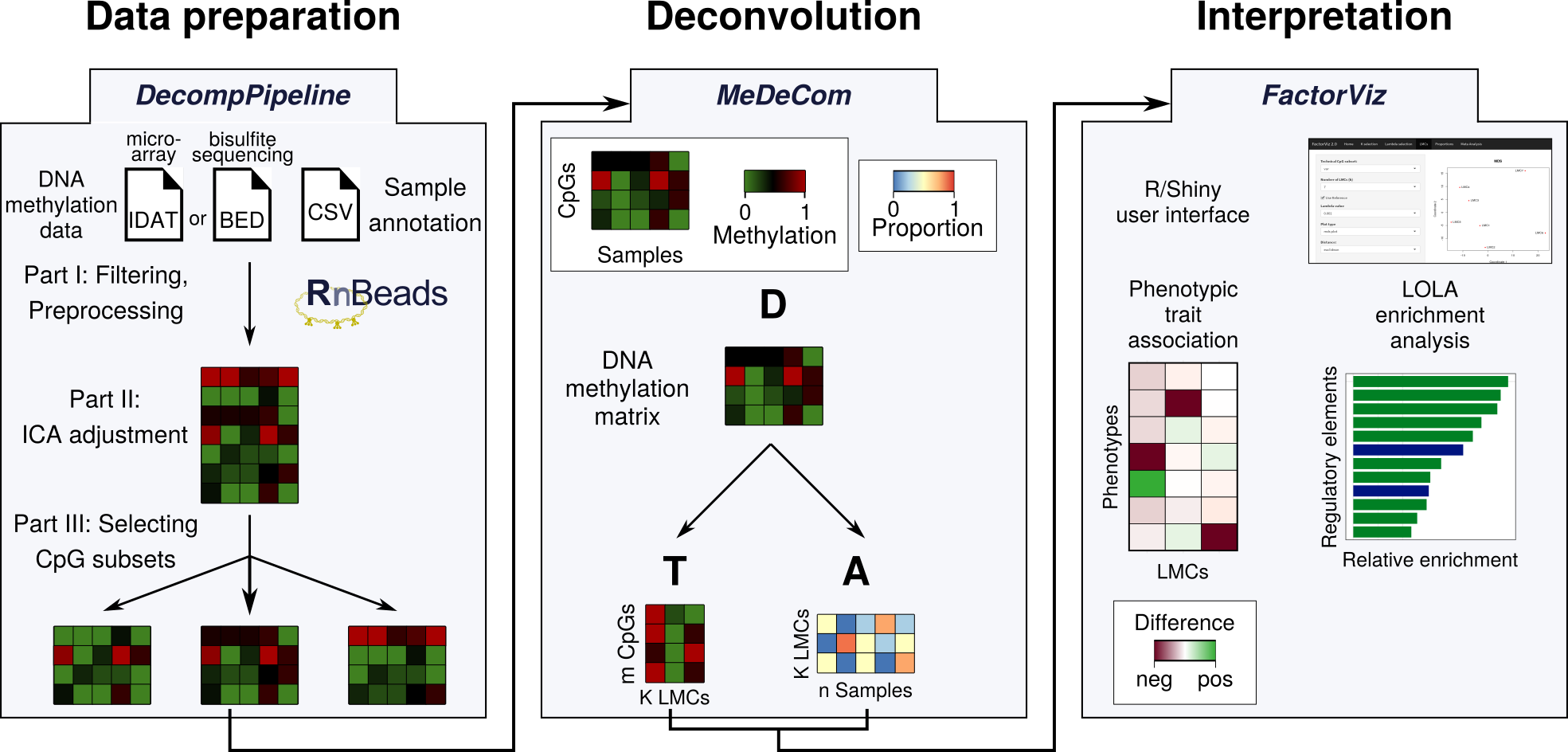
Reference-free deconvolution, visualization and interpretation of complex DNA methylation data using DecompPipeline, MeDeCom and FactorViz
DNA methylation assays are typically performed on bulk tissue samples, which show substantial cellular heterogeneity. To dissect these heterogeneous DNA methylomes, several deconvolution approaches have been proposed (for a list of tools see this GitHub page). Reference-free deconvolution tools do not rely on purified cell type profiles, and address heterogeneity in an unbiased way. These methods rely on high-quality data and pose challenges for the interpretation of deconvolution results. We propse a three-stage protocol to perform reference-free deconvolution of complex DNA methylation data:
Data preparation
Input DNA methylation data either obtained from bisulfite sequencing technologies (BED files) or the Illumina Infinium BeadArrays (IDAT files) are processed using the RnBeads sofware package. The data set is checked for quality using RnBeads' reporting functionality, and then filtered according to user-specified options, which removes intensity/coverage outliers, or annotated single nucleotide polymorphisms. These steps are compiled in the comprehensive software suite DecompPipeline. Confounding factors are addressed for using Independent Component Analysis (ICA).
Deconvolution
The second stage comprises reference-free deconvolution of the processed DNA methylation data matrix. The protocol supports MeDeCom, as well as RefFreeCellMix and EDec. All of these methods are based on non-negative matrix factoriation (NMF).
Interpretation
To visualize deconvolution results and to get from the results to biological interpretation, we implemented the R/shiny graphical user interface FactorViz. FactorViz provides functions to link the obtained components (latent methylation components=LMCs) to phenotypic traits and to perform enrichment analysis on the sites that are particularly hypo-methylated in an LMC.
Publication
The protocol has been published in Nature Protocols, 2020
Developers
The protocol has been mainly developed by Pavlo Lutsik and Michael Scherer in close collaboration with Petr Nazarov. Further contributions have been made by Reka Toth, Shashwat Sahay, Valentin Maurer, Tony Kaoma, Nikita Vedeneev, Christoph Plass, Thomas Lengauer, and Jörn Walter. The developers hold positions at the Department of Genetics/Epigenetics at Saarland University, at the Divsion of Cancer Epigenomics at the German Cancer Research Center, at the Quantitative Biology Unit at Luxembourg Insitute of Health and at the Research Group Computational Biology at the Max Planck Insitute for Informatics. The project is supported by the German Epigenome Program, de.NBI-epi, and SYSCID.






